 |
Module Overview
MicrobiomeAnalyst is a user-friendly web-based platform developed to enable comprehensive statistics, visualization,
functional interpretation, and integrative analysis of common datasets from microbiome studies based on updated methods and databases.
The current MicrobiomeAnalyst (2.0) supports raw sequence processing, statistical analysis, functional prediction,
and meta-analysis for marker gene data, multiple approaches for shotgun data profiling, taxon set enrichment analysis and
integrative analysis of microbiome and metabolomics data.
|
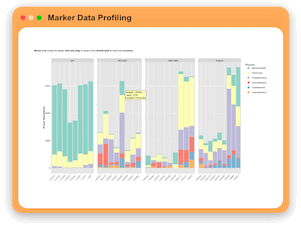 |
Marker Data Profiling
MicrobiomeAnalyst allows users to perform different types of analyses on maker gene count table including: visual exploration through
interactive stack barplot and pie chart, rarefaction curve and phylogenetic tree, community profiling through diversity analysis,
clustering and correlation through interactive heatmaps, dendrogram and correlation network, comparison and classification
through multi-factor comparision analysis, LEfSe and Random Forest, as well as functional prediction through PICRUSt, Tax4Fun and Tax4Fun2.
|
 |
Shotgun Data Profiling
MicrobiomeAnalyst provides multiple analytical approaches for the analysis of both KO count table and KO list including: functional profiling through diversity overview and association analysis, clustering analysis through
interactive heatmap, dendrogram and PCA visualization, multi-factor comparision analysis as well as biomarker analysis using the methods of LEfSe and Random Forest.
|
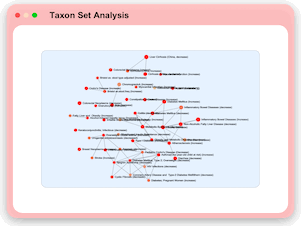 |
Taxon Set Analysis
MicrobiomeAnalyst now allows users to identify taxonomic signatures characterized by their shared functions or associations with different phenotypes through enrichment analysis against manually curated taxon set libraries including:
700+ SNP associated taxon sets, 500 environment associated taxon sets, 454 disease associated taxon sets, 221 diet and lifestyle associated taxon sets, 137 drugs and antibiotic associated taxon sets, 102 host immune responses associated taxon sets, etc.
|
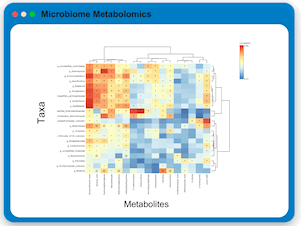 |
Microbiome Metabolomics Profiling
Users can upload paired microbiome and metabolomics data to perform integrative analysis using statistical-driven and knowledge-driven approaches.
MicrobiomeAnalyst currently offers three strategies: dimensionality reduction, contextualized metabolic network analysis, and
knowledge-enhanced microbiome-metabolome correlation analysis. The results can be viewed in 3D scatter plot, interactive metabolic network
and interactive heatmap for intuitive visual exploration and understanding.
|
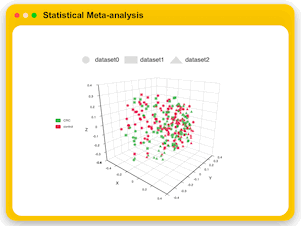 |
Statistical Meta-analysis
Users can upload multiple marker gene data sets collected under comparable conditions to identify shared biomarkers across multiple studies.
MicrobiomeAnalyst currently offer three analysis tracks for user to explore the consistent pattern and potential biomarkers,
which include visual exploration through stacked bar/area plot or PCoA plots, alpha and bete diversity meta-analysis, and biomarker meta-analysis.
|
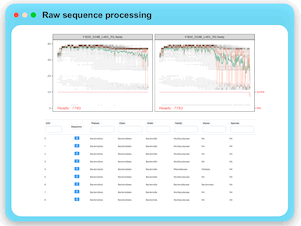 |
Raw Data Processing
MicrobiomeAnalyst now supports the raw sequence processing for marker gene data (16S/18S/ITS). User can upload their raw reads together
with a metadata file for further processing. The implementation is based on the well-established DADA2 pipeline. An ASV abundance table
and a taxonomy annotation table are provided for download or directly submit for downstream statistical analysis.
|
 |
Report Generation & Batch Processing
Upon completion of your analysis, MicrobiomeAnalyst will generate a comprehensive PDF report documenting each step performed along with
corresponding tabular and graphical results. All processed data and images are also freely available for download.
Users can also download the R history and install the underlying MicrobiomeAnalystR package for batch processing and reproducible analysis.
|
|







